From the Past to Modern Detection
Sepsis has been a medical challenge for centuries. The term "sepsis" is derived from the Greek word σῆψις, meaning decay or putrefaction, indicating the disease's destructive nature. The earliest recorded observations of sepsis date back to the writings of Hippocrates (circa 460–370 BC), who described the process of wound putrefaction and the systemic illness that followed. But for thousands of years, our basic knowledge of sepsis was limited, and many wrong ideas about it continued to exist.
Current Methods of Detecting Sepsis in Hospitals
Today's hospitals use advanced clinical criteria, laboratory testing, and technology to detect sepsis early. The implementation of SOFA and qSOFA scores aids in rapid assessment, while biomarkers like Procalcitonin and C-reactive protein provide key diagnostic insights. Point-of-care testing technologies enable bedside diagnostics, offering quicker intervention possibilities. Electronic Health Record (EHR) systems with integrated sepsis detection algorithms play a crucial role in identifying sepsis signs early, and antimicrobial stewardship programs ensure effective and responsible use of antibiotics to treat sepsis.
The evolution from historical misconceptions to current evidence-based practices highlights the significant strides made in sepsis detection and management, emphasizing the critical role of early detection and timely, appropriate treatment in improving patient outcomes.
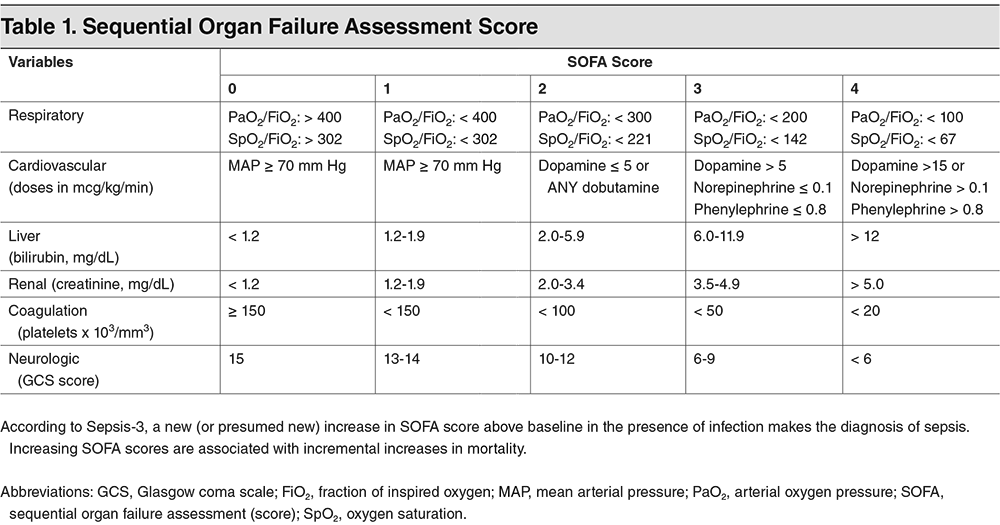
source: https://www.ebmedicine.net/topics/infectious-disease/sepsis-septic-shock